

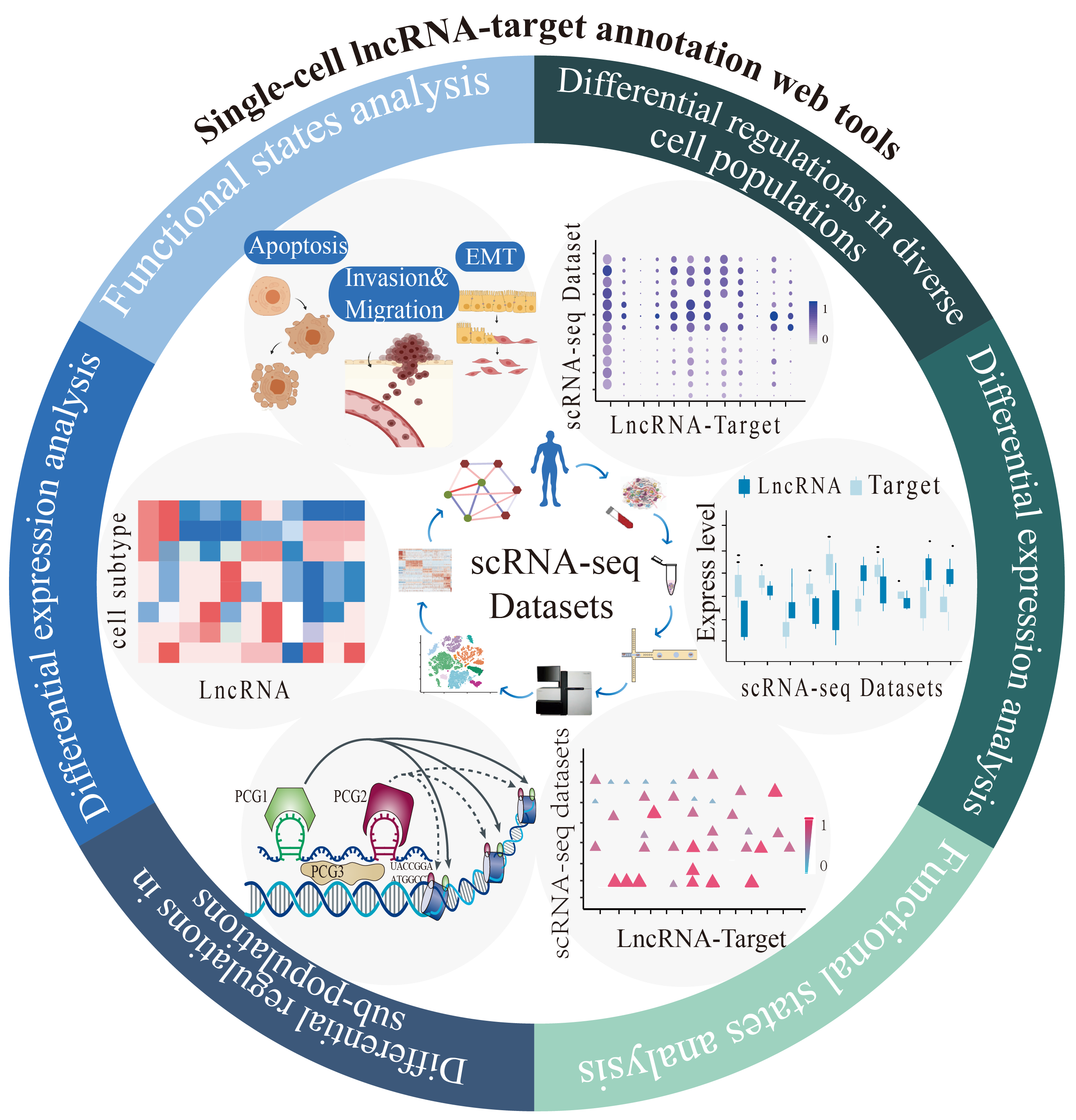
The Single Cell Web tool provides single-cell lncRNA-target annotation based on 31 single-cell RNA sequencing
(scRNA-seq) datasets related to 21 human cancers (>500 cells). It provides interactive and customizable functions
including two powerful modules:
(1) pan-cancer analysis module: graphically display the differences/similarities of lncRNA-target
regulations among cancer types at the single-cell level.
(2) cancer-specific analysis module: reveal the cancer cell sub-populations specificity of lncRNA-target
regulations and intra-tumor heterogeneity at the single-cell level.
In summary, from the perspective of pan-cancer and cancer-specific analysis, Single Cell Web Tools characterize
lncRNA-target regulations from scRNA-seq datasets, including differential lncRNA-target regulations analysis,
differential expression analysis, relevant functional states analysis, lncRNA-target networks, cell clustering maps,
distribution in diverse cell populations and in sub-populations. View our Help page for further help.
This function provides the differential analysis of different lncRNA-target regulation activity scores in 31 single-cell datasets containing 21 human diseases. Differential lncRNA-target regulation in a given single-cell dataset was identified by comparison with other single-cell datasets.
This function provides the landscape of differential expression of lncRNAs and target genes across 31 single-cell datasets with 21 human cancers.
This function provides the landscape of differential lncRNA-target regulations associated with a given cell state across 31 single-cell datasets with 21 human cancers. Differential lncRNA-target regulations with a given cell state in a given single-cell data set were identified by comparing with other single-cell data sets.
This function provides the differences in the distribution of lncRNA-target regulation activity scores among cancer cell sub-populations. It will help to understand the roles of lncRNA-target regulations in intra-tumor heterogeneity at the single-cell level.
This function provides the differential expression of lncRNAs and target genes across cancer cell sub-populations.
This function provides the activity scores of differential lncRNA-target regulations associated with a given cell state among cancer cell sub-populations in a single-cell data sets.
| ID | Cancer Type | Cell Number | lncRNA-Target number | Source | Accession | Cell Type |
| (UM)GSE139829 | Uveal melanoma | 42230 | 2040 | Tissue: Eye | GSE139829 | Neurons,Tissue_stem_cells,T_cells,Macrophage |
| (NSCLC)GSE143423_Met1 | Non-small Cell Lung Cancer | 13382 | 952 | Tissue: Brain | GSE143423 | Epithelial_cells,Macrophage,NK_cell |
| (UCEC)GSE139555_Uterus | Endometrial adenocarcinoma | 12758 | 48 | Tissue: Endometrium | GSE139555 | Pre-B_cell_CD34-,Epithelial_cells,T_cells |
| (CRC)GSE139555_Colon | Colorectal cancer | 10112 | 378 | Tissue: Colon | GSE139555 | B_cell,T_cells,NK_cell,CMP,Monocyte,Neurons,Chondrocytes |
| (NSCLC)GSE143423_Met2 | Non-small Cell Lung Cancer | 9595 | 804 | Tissue: Brain | GSE143423 | Epithelial_cells,Macrophage,Endothelial_cells,Tissue_stem_cells,Astrocyte |
| (PAAD)GSE112845 | Pancreatic cancer | 8321 | 1198 | Cell Line: KLM1 | GSE112845 | MT-ND2+ Epithelial_cells,FOXM1+ Epithelial_cells,LRRC75A+ Epithelial_cells,SPP1+ Epithelial_cells,NUPR1+ Epithelial_cells,DHRS2+ Epithelial_cells,UPP1+ Epithelial_cells,KRTAP3-1+ Epithelial_cells,UBE2T+ Epithelial_cells |
| (PDAC)PRJCA001063_IB | Pancreatic Ductal Adenocarcinoma | 8202 | 804 | Tissue: Head | PRJCA001063 | Tissue_stem_cells,Endothelial_cells,T_cells,Monocyte,B_cell,Epithelial_cells |
| (ARMS)GSE113660 | Alveolar rhabdomyosarcoma | 6875 | 887 | Cell Line: Rh41 | GSE113660 | Neurons,MSC |
| (PDAC)PRJCA001063_IIA | Pancreatic Ductal Adenocarcinoma | 6289 | 958 | Tissue: Head | PRJCA001063 | Epithelial_cells,Monocyte,Tissue_stem_cells,Chondrocytes,Pre-B_cell_CD34-,T_cells |
| (HNSC)GSE103322 | Head and neck cancer | 5902 | 2151 | Tissue: Lymph node | GSE103322 | Monocyte,T_cells,Endothelial_cells,Epithelial_cells,Fibroblasts,Chondrocytes,Tissue_stem_cells,Astrocyte,B_cell |
| (ATC)GSE148673 | Anaplastic Thyroid Cancer | 5861 | 583 | Tissue: Fresh tumor | GSE148673 | T_cells,B_cell,NK_cell,Monocyte,Fibroblasts |
| (OV)E-MTAB-8559_IIIB | Ovarian Carcinoma | 5846 | 1507 | Tissue: Peritoneal fluid | E-MTAB-8559 | Smooth_muscle_cells,Neurons,Fibroblasts,Tissue_stem_cells,MSC,Endothelial_cells |
| (MPAL)GSE139369 | Mixed-phenotype Acute Leukemia | 5643 | 385 | Tissue: Bone Marrow | GSE139369 | Monocyte,CMP,Platelets |
| (PDAC)PRJCA001063_IIB | Pancreatic Ductal Adenocarcinoma | 5263 | 1046 | Tissue: Body and tail | PRJCA001063 | Chondrocytes,T_cells,Macrophage,Monocyte,Epithelial_cells,Fibroblasts,Tissue_stem_cells,NK_cell,Smooth_muscle_cells,Endothelial_cells |
| (NSCLC)GSE143423_Met3 | Non-small Cell Lung Cancer | 5169 | 1350 | Tissue: Brain | GSE143423 | Macrophage,Tissue_stem_cells,Epithelial_cells,Monocyte,T_cells,NK_cell,Astrocyte |
| (OV)E-MTAB-8559_IIIC | Ovarian Carcinoma | 5046 | 1588 | Tissue: Peritoneal fluid | E-MTAB-8559 | MSC,Smooth_muscle_cells,Fibroblasts,Tissue_stem_cells,Embryonic_stem_cells,Epithelial_cells |
| (SARC)GSE146221_CHLA9 | Ewing sarcoma | 4865 | 1421 | Cell Line: CHLA9 | GSE146221 | AL844908.1+ Neurons,AC103706.1+ Neurons,TAGLN+ Neurons,PIF1+ Neurons,FAM111B+ Neurons,HDAC10+ Neurons |
| (SKCM)GSE72056 | Melanoma | 4645 | 2021 | Tissue: Skin | GSE72056 | B_cell,T_cells,Epithelial_cells,Neurons,Tissue_stem_cells,Endothelial_cells,Monocyte |
| (SARC)GSE146221_CHLA10 | Ewing sarcoma | 4632 | 1033 | Cell Line: CHLA10 | GSE146221 | Neuroepithelial_cell,iPS_cells,MSC,Neurons |
| (BLCA)GSE130001 | Bladder cancer | 4129 | 718 | Tissue: Bladder | GSE130001 | Endothelial_cells,Epithelial_cells,Keratinocytes,Chondrocytes,Tissue_stem_cells |
| (CHOL)GSE125449 | Intrahepatic cholangiocarcinoma | 3835 | 703 | Tissue: Liver | GSE125449 | T_cells,Monocyte,Endothelial_cells,Hepatocytes,B_cell,Tissue_stem_cells |
| (SARC)GSE146221_TC71 | Ewing sarcoma | 3673 | 1240 | Cell Line: TC71 | GSE146221 | Neurons,iPS_cells |
| (NSCLC)E-MTAB-6149 | Non-small cell lung cancer | 3524 | 910 | Tissue: Lung | E-MTAB-6149 | Epithelial_cells,Embryonic_stem_cells,T_cells,iPS_cells |
| (NET)GSE140312 | Gastrointestinal Neuroendocrine Cancer | 3158 | 364 | Tissue: Liver | GSE140312 | Endothelial_cells,Fibroblasts,Hepatocytes,Neurons,Monocyte |
| (HGG)GSE103224 | High-grade glioma | 2772 | 1035 | Tissue: Brain | GSE103224 | Neuroepithelial_cell,Chondrocytes,Astrocyte |
| (OV)GSE118828 | Ovarian cancer | 1909 | 945 | Tissue: Ovary | GSE118828 | NK_cell,Epithelial_cells,Monocyte,Tissue_stem_cells,Fibroblasts |
| (CML)GSE76312-Tissue Bone marrow | Chronic myelogenous leukemia | 1877 | 1908 | Tissue: Bone marrow | GSE76312 | CMP,Pro-B_cell_CD34+ |
| (AML)GSE110499 | Acute myeloid leukemia | 1184 | 592 | Tissue: Bone marrow | GSE110499 | B_cell,Monocyte,T_cells,MEP |
| (GBM)GSE84465 | Glioblastoma | 991 | 2099 | Tissue: Brain | GSE84465 | Astrocyte,Tissue_stem_cells |
| (GBM)GSE57872-Tissue Brain | Glioblastoma | 623 | 2010 | Tissue: Brain | GSE57872 | ZMAT5+ Astrocyte,OLIG2+ Astrocyte,TOP2A+ Astrocyte,LMX1B+ Astrocyte,SMYD2+ Astrocyte,SAA2+ Astrocyte,NNAT+ Astrocyte |
| (MEL)GSE99330 | Melanoma | 575 | 1669 | Cell Line: WM989 | GSE99330 | Neurons,T_cells |
| (BRCA)GSE77308 | Breast cancer | 369 | 2712 | PDX | GSE77308 | MSC,Epithelial_cells |
| (BRCA)GSE75688 | Breast cancer | 317 | 2885 | Tissue: Breast | GSE75688 | Epithelial_cells,B_cell |
| (ESCC)GSE81812 | Esophageal squamous cell carcinoma | 314 | 2818 | Cell Line: KYSE-180 | GSE81812 | RSPH10B+ Epithelial_cells,CYP24A1+ Epithelial_cells,LINC00954+ Epithelial_cells,CTA-292E10.8+ Epithelial_cells,PIF1+ Epithelial_cells |
| (MEL)GSE81383 | Melanoma | 307 | 2403 | Tissue: Skin | GSE81383 | iPS_cells,Neurons |
| (CRC)GSE81861-Tissue Colon | Colorectal cancer | 290 | 1639 | Tissue: Colon | GSE81861 | PDCD11+ Epithelial_cells,SLITRK6+ Epithelial_cells,TNFRSF11B+ Epithelial_cells,CENPQ+ Epithelial_cells |
| (CML)GSE98734 | Chronic myelogenous leukemia | 267 | 2273 | Cell Line: K562 | GSE98734 | CTH+ BM & Prog.,USO1+ BM & Prog.,CCNO+ BM & Prog.,RP11-474I11.8+ BM & Prog.,ATP8B2+ BM & Prog.,BAHD1+ BM & Prog. |
| (LUAD)DRP001358 | Lung adenocarcinoma | 245 | 2660 | Cell Line: LC-2/ad | DRP001358 | H19+ Epithelial_cells,HMMR+ Epithelial_cells,AGAP1-IT1+ Epithelial_cells,C8orf47+ Epithelial_cells |
| (LUAD)DRP003337 | Lung adenocarcinoma | 218 | 1923 | Cell Line: A549 | DRP003337 | Epithelial_cells,Smooth_muscle_cells |
| (GBM)GSE57872 | Glioblastoma | 177 | 3487 | Cell Line: GSC/DGC | GSE57872 | LA16c-366D1.3+ Astrocyte,UBB+ Astrocyte,LSM11+ Astrocyte,CYTL1+ Astrocyte |
| (NSCLC)DRP003981-Cell Line PC-9 | Non-small cell lung cancer | 164 | 2661 | Cell Line: PC-9 | DRP003981 | RAB3IL1+ Epithelial_cells,NUPR1+ Epithelial_cells,CEP152+ Epithelial_cells |
| (PC)GSE99795 | Prostate cancer | 144 | 2381 | Cell Line: LNCaP | GSE99795 | iPS_cells,Epithelial_cells |
| (CCA)ERP020478 | Cervix cancer | 126 | 3360 | Cell Line: Hela | ERP020478 | MSC,Epithelial_cells |
| (LUAD)GSE69405-PDX | Lung adenocarcinoma | 126 | 2458 | PDX | GSE69405 | Epithelial_cells,Endothelial_cells |
| (CRC)GSE51254 | Colorectal cancer | 99 | 2271 | Cell Line: HCT-116 | GSE51254 | Epithelial_cells,iPS_cells |
| (BCA)DRP003981 | Bronchoalveolar carcinoma | 96 | 1872 | Cell Line: H1650 | DRP003981 | Epithelial_cells |
| (NSCLC)DRP003981-Cell Line H1975 | Non-small cell lung cancer | 96 | 1929 | Cell Line: H1975 | DRP003981 | TFPI2+ Epithelial_cells,HLA-DPA1+ Epithelial_cells |
| (NSCLC)DRP003981-Cell Line H2228 | Non-small cell lung cancer | 96 | 1967 | Cell Line: H2228 | DRP003981 | RACGAP1+ Epithelial_cells,DYNLL1+ Epithelial_cells |
| (LUAD)DRP003981-Cell Line II18 | Lung adenocarcinoma | 96 | 2302 | Cell Line: II18 | DRP003981 | S100A9+ Epithelial_cells,DGKA+ Epithelial_cells,CTD-2636A23.2+ Epithelial_cells |
| (MLPS)E-MTAB-6142 | Myxoid liposarcoma | 96 | 1769 | Cell Line: MLS 1765-92 | E-MTAB-6142 | AURKA+ MSC,RIBC2+ MSC |
| (PDAC)GSE99305 | Pancreatic ductal adenocarcinoma | 96 | 1998 | Cell Line: Pancreatic ductal adenocarcinoma cell lines | GSE99305 | NUF2+ Epithelial_cells,TP53INP2+ Epithelial_cells |
| (CML)GSE76312 | Chronic myelogenous leukemia | 91 | 1714 | Cell Line: K562 | GSE76312 | LRRC16A+ BM & Prog.,AHSP+ BM & Prog. |
| (MEL)GSE97681-Cell Line WM983B | Melanoma | 86 | 3358 | Cell Line: WM983B | GSE97681 | CTB-151G24.1+ Neurons,RP11-542A14.2+ Neurons,POSTN+ Neurons |
| (RCC)GSE73121 | Renal cell carcinoma | 83 | 2403 | PDX | GSE73121 | PAEP+ Epithelial_cells,G0S2+ Epithelial_cells |
| (LUAD)GSE81861-Cell Line A549 | Lung adenocarcinoma | 74 | 1422 | Cell Line: A549 | GSE81861 | Epithelial_cells |
| (CML)GSE81861 | Chronic myelogenous leukemia | 73 | 1831 | Cell Line: K562 | GSE81861 | BM & Prog.,MEP |
| (BRCA)GSE75367 | Breast cancer | 70 | 2389 | CTC | GSE75367 | Epithelial_cells,Platelets |
| (HCC)GSE103866 | Hepatocellular carcinoma | 55 | 2031 | Cell Line: HuH-7 | GSE103866 | iPS_cells |
| (CRC)GSE81861-Colorectal cancer | Colorectal cancer | 51 | 1679 | Cell Line: HCT-116 | GSE81861 | Embryonic_stem_cells |
| (CML)GSE69405 | Chronic myelogenous leukemia | 50 | 2168 | Cell Line: K562 | GSE94979 | Epithelial_cells |
| (LUAD)GSE94979 | Lung adenocarcinoma | 50 | 1081 | Cell Line: H358 | GSE69405 | BM & Prog. |
| (NSCLC)GSE81861-Cell Line H1437 | Non-small cell lung cancer | 47 | 1238 | Cell Line: H1437 | GSE81861 | Neuroepithelial_cell,BM & Prog. |
| (LUAD)DRP001358-Cell Line PC-9 | Lung adenocarcinoma | 46 | 2411 | Cell Line: PC-9 | DRP001358 | Epithelial_cells |
| (LUAD)DRP001358-Cell Line VMRC-LCD | Lung adenocarcinoma | 46 | 2008 | Cell Line: VMRC-LCD | DRP001358 | iPS_cells |
| (BRCA)GSE91395 | Breast cancer | 45 | 422 | Cell Line: MCF-7 | GSE91395 | HMGB1+ Epithelial_cells,DHRS2+ Epithelial_cells |
| (HCC)GSE103866-Cell Line HuH-1 | Hepatocellular carcinoma | 43 | 1927 | Cell Line: HuH-1 | GSE103866 | Hepatocytes |
| (NB)GSE90683 | Neuroblastoma | 36 | 3014 | Cell Line: Neuroblastoma cell line | GSE90683 | Neurons |
| (MEL)GSE97681-Cell Line WM989 | Melanoma | 36 | 3289 | Cell Line: WM989 | GSE97681 | Neurons,MSC |
| (RCC)GSE73121-Tissue Kidney | Renal cell carcinoma | 35 | 2050 | Tissue: Kidney | GSE73121 | Epithelial_cells |
| (BRCA)GSE91395-Cell Line MDA-MB-231 | Breast cancer | 33 | 374 | Cell Line: MDA-MB-231 | GSE91395 | MSC |
| (HCC)GSE65364 | Hepatocellular carcinoma | 26 | 778 | Tissue: Liver | GSE65364 | Hepatocytes |
| (AML)E-MTAB-2672 | Acute myeloid leukemia | 24 | 2587 | Tissue: Bone marrow | E-MTAB-2672 | GMP |
| (ALL)GSE100694 | Acute lymphoblastic leukemia | 24 | 3394 | Cell Line: LOUCY/JURKAT | GSE100694 | Pro-B_cell_CD34+ |
This function provides the landscape of the proportion of cells with high activity scores (> 0.5*max (AUC scores)) for differential lncRNA-target regulations in cells across 31 single-cell datasets with 21 human cancers. Wilcoxon rank sum test was used to identify differential lncRNA-target regulations by comparing the activity scores between a given scRNA-seq and other scRNA-seq datasets.
scRNA-seq Dataset: Select a cancer single-cell data set to obtain differential lncRNA-target regulations.
LncRNA/Gene Symbol: Select a lncRNA to obtain lncRNA-target regulations for analysis.
This function provides the landscape of differential expression of lncRNAs and target genes across 31 single-cell datasets with 21 human cancers. Wilcoxon rank sum test was used to identify differential lncRNA and genes by comparing expression levels between a given scRNA-seq and other scRNA-seq datasets.
scRNA-seq Dataset: Select a cancer single-cell data set for differential expression analysis.
LncRNA/Gene Symbol: Select a lncRNA for differential expression analysis.
This function provides the landscape of the proportion of cells with high activity scores (> 0.5*max (AUC scores)) for differential lncRNA-target regulations associated with a given cell state (such as stemness, epithelial-mesenchymal transition, angiogenesis and inflammation) across 31 single-cell datasets with 21 human cancers. Wilcoxon rank sum test was used to identify differential lncRNA-target regulations by comparing the activity scores between a given cell functional state and other cell functional states.
scRNA-seq Dataset: Select a cancer single-cell data set for differential regulation analysis.
Cell functional states: Select a cancer single-cell data set for differential regulation analysis.
LncRNA/Gene Symbol: Select a lncRNA for drawing the point diagram.
This function provides the differences in the distribution of lncRNA-target regulation activity scores among cancer cell sub-populations and shows the proportion of cells with high activity scores (> 0.5*max (AUC scores)). It will help to understand the roles of lncRNA-target regulations in intra-tumor heterogeneity at the single-cell level. Wilcoxon rank sum test was used to identify differential lncRNA-target regulations by comparing the activity scores between a given cancer cell sub-population and other cancer cell sub-populations.
scRNA-seq Dataset: Select a cancer single-cell data set for analysis.
Cancer cell sub-populations: Select a cancer cell sub-population for differential regulations analysis.
LncRNA/Gene Symbol: Select a lncRNA to obtain key lncRNA-target regulations for analysis.
This function provides the differential expression of lncRNAs and target genes across cancer cell sub-populations. Wilcoxon rank sum test was used to analyze the differential expression of lncRNA and its target genes in different cancer cell sub-populations.
LncRNA: Select a lncRNA for differential expression analysis.
scRNA-seq Dataset: Select a cancer single-cell data set for analysis.
Cancer cell sub-populations: Select a cancer cell sub-population for differential regulations analysis.
LncRNA/Gene Symbol: Select a lncRNA for differential expression analysis.
This function provides the cell proportion with high activity scores (> 0.5*max (AUC scores)) of differential lncRNA-target regulations associated with a given cell state (such as stemness, epithelial-mesenchymal transition, angiogenesis and inflammation) among cancer cell sub-populations in a single-cell data sets. Wilcoxon rank sum test was used to identify differential lncRNA-target regulations by comparing the activity scores between a given cell functional state and other cell functional states.
scRNA-seq Dataset: Select a cancer single-cell data set for analysis.
Cell functional states: Select a cell functional state for analysis.
LncRNA/Gene Symbol: Select a lncRNA to obtain key lncRNA-target regulations for analysis.
Copyright © 2019 College of Bioinformatics Science and Technology, Harbin Medical University All rights reserved.
黑ICP备2022004059号